|
Introduction
Cells internalization of proteins is regulated by membrane protein-protein interactions. Inside cells, the STAM2 protein encodes the transcription signal of endocytosis. Ubiquitin allows the recognition of targeted proteins to degrade, fixing on its surface. Ubiquitination is a reversible process: desubiquitination is catalyzed by the AMSH protein. Here, the interaction between the UIM-SH3 domain of STAM2 and a 14 amino acids peptide of AMSH is characterized. An NMR titration method of chemical shift mapping of surface interactions was used. Our results suggest a strong interaction between AMSH peptide and the SH3 domain of STAM2, with a dissociated constant of 10 ± 6 µmol/L.
Experimental conditions
STAM2 (UIM-SH3) was produced with a [1H-15N] isotopic labeling, expressed with a NusA-hexahistidine tag on the N-terminal and then purified by Nickel affinity chromatography and gel-filtration. The AMSH peptide was obtained from the compagny Genosphere. Samples were prepared in a 20 mmol/L phosphate buffer at pH 6,8, with 0,02% (v/v) of NaN3, 10% (v/v) of DO and an initial protein concentration of 180 µmol/L. All NMR experiments were acquired with a Varian Inova 600 MHz spectrometer. UIM-SH3 residues assignment was obtained by the following 3D NMR methods: HNCA, HN(CO)CA, HNCO and HN(CO)CACB. The titration of UIM-SH3 by AMSH peptide was realized at 310 K, following residues chemical shift perturbations; by superposition of a series of [1H-15N]-HSQC spectrum and adding AMSH peptide until a 2,8 ratio of [AMSH peptide] / [UIM-SH3]. Data was transformed with NMR Pipe, and analyzed with SPARKY and MATLAB.
Results
According to the NMR HSQC chemical shift mapping presented on figure 1(A), thirteen residues are in interaction with AMSH peptide. Those amino acids involved in the binding are localized on the SH3 domain of STAM2. They are characterized by a total chemical shift of 0,08 to 0,28 ppm. Figure 1(B) shows the bar chart of total chemical shift variation for each amino acid of UIM-SH3. As depicted by the 3D structure of UIM-SH3 on figure 2, residues involved in the interaction are hydrophobic amino acids of the SH3 domain: S260 situated on the 310 helix, L257 on a β-sheet, D242, S242, N245 on the n-Src loop and Y218, D219, E221, A222, V223, E227, L228 on the RT loop. The dissociation constant (KD) associated to this binding is about 10 ± 6 µmol/L. In physiologic conditions of 310 K at pH 6,8 , our essay suggests a strong specific interaction between the SH3 domain of STAM2 and AMSH.
Conclusion
SH3 domain is known to interact with proteins with proline-rich protein. Indeed, our results demonstrate a strong affinity between this domain and the AMSH protein. Those results indicate the capital role of AMSH to catalyze the regulator desubiquitination process.
|
|
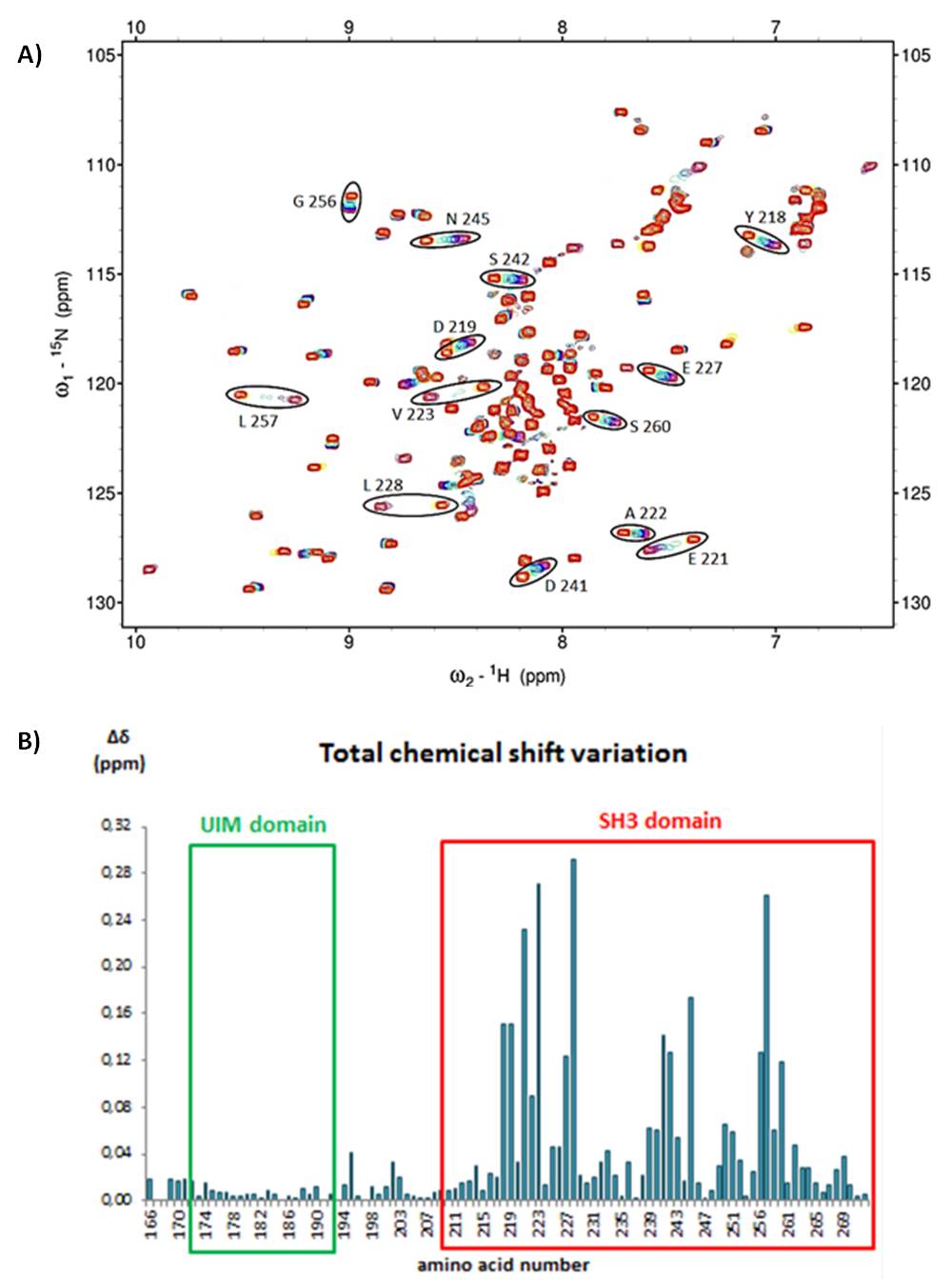
A) NMR superposed [1H15N]-HSQC of titration of UIM-SH3 by AMSH peptide. Residues with a chemical shift modified are circled in black.
B) Bar chart of total chemical shift variation for each amino acid of UIM-SH3.
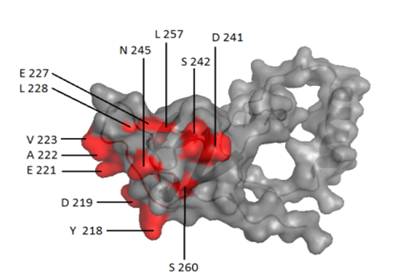
3D structure of UIM-SH3’s surface of binding with AMSH peptide. Concerned residues are colored in red.
|




