|
Introduction
Mass spectrometry is an analytical technique, which generates ions separated by their mass-to-charge ratio (m/z). Currently, this technique is notably applied to characterize and quantify macromolecules such as proteins for system biology or in clinical research. In the ANABIO-MS laboratory (Analyses of Biomolecules by Mass Spectrometry) of the Institute of Analytical Sciences, a quantification method based on liquid chromatography coupled to tandem mass spectrometry (MRM mode) was developed, targeting 700 proteins to study the proteome of a clinical Staphylococcus aureus strain.
Experimental conditions
Cell pellets of two inactivated Staphylococcus aureus strains were used for the experiment. After extraction, proteins were digested by an endopeptidase (trypsin) to generate tryptic peptides. Trypsin cleaves peptide bonds of the basic amino acid residues lysin and arginin. To simplify the peptide mixture, a MCX solid phase extraction (MCX refers to a cation-exchanged phase) was performed and four fractions of eluted peptides were collected. Each fraction was concentrated under nitrogen flow and finally, peptides were recovered with a water/methanol solvent mixture.
Then, a shotgun proteomic analysis was performed on a first Staphylococcus aureus strain (strain 1) with a high resolution mass spectrometer (Q-Exactive Orbitrap). The full MS scan range was fixed at 400 to 1200 m/z with a resolution of 35,000 at 200 m/z. The resulting data were compared with a proteomic database (UniProt) to attribute each fragmentation spectrum to a Staphylococcus aureus peptide. Next, one or two peptides were picked up for each identified protein with three transitions retained per peptide. The final MRM assay gathered 1055 peptides surrogates of 777 proteins monitored with 3215 transitions. At last, a MRM analysis was performed on a QTrap 6500 mass spectrometer to test the method on a secondary Staphylococcus aureus strain (strain 2).
Results
MRM data were processed with a “Skyline” platform with which relative information such as peptide retention times, peak areas and fragment intensities can be visualized as graphs and chromatograms (Figure 1). From this analysis, 65.5% of proteins were detected; for proteins characterized in the multiplex method by two peptides, 80.6% were found (but only 57.1% with one) (Figure 2). Thus, it is better to pick up two peptides for each protein instead of one.
For the non-detected proteins, the main hypothesis is that some proteins could be expressed in the first strain and not in the second. Another possibility could be that some peptide transitions were too weak to be distinguished from the background noise.
Conclusion
With the developed multiplex method on Staphylococcus aureus on Strain 1, the analysis on the second strain reveals that 65.5% of all proteins have been detected. Among the rest (34.5%), some proteins might not be expressed in this strain or were not detected due to weaker signal-to-noise ratio. Moreover, 80.6% of proteins monitored with two peptides were detected (for only 57.1% for proteins with one). In the near future, new proteins, either from in-house discovery proteomic data or from publicly available proteomic repositories, will still enrich the MRM assay.
|
|
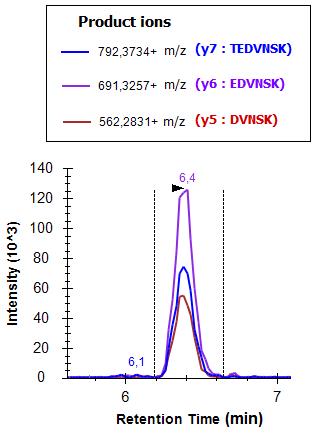
Chromatogram showing the three transitions of a peptide (LTEDVNSK corresponding to a m/z precursor value of 453.2324++) of Staphylococcus aureus, strain 2. The ‘+’ number refers to the positive charge of the precursor or peptide ion.
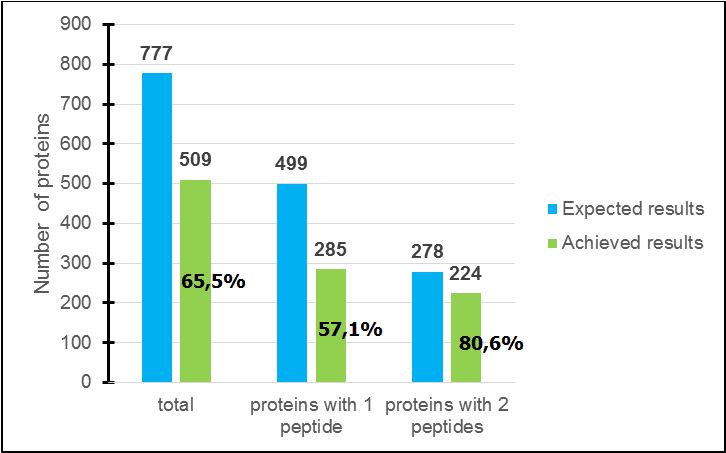
Comparison between expected and achieved results relative to proteins characterized with one or two peptides in the multiplex method.
|




